A once-forgotten antibiotic could be a new weapon against drug-resistant infections
Doctors may have a new tool to protect patients against multi-drug resistant bacterial infections. But the new defense against increasingly difficult-to-treat bacteria isn’t a brand new development—it is an 80 year-old antibiotic. A study published May 16 in the open access journal PLOS Biology looked at a natural product made in soil called nourseothricin that was discovered in 1942.
The rise of antibiotic-resistant bacterial infections has encouraged a search for new antibiotics. Antibiotic resistance is a very serious and growing medical problem—according to the Centers for Disease Control and Prevention, antimicrobial resistance killed at least 1.27 million people worldwide and was associated with close to 5 million deaths in 2019. In the United States, more than 2.8 million antimicrobial-resistant infections occur each year, and over 35,000 people die as a result.
Nourseothricin contains multiple forms of a complex molecule called streptothricin. There were high hopes that the streptothricin inside would be a powerful agent against bacteria called gram-negative bacteria. These bacteria, such as E.coli., have a thick outer protective layer and are particularly hard for antibiotics to kill.
Unfortunately, nourseothricin was toxic to kidneys according to the results of an unpublished limited human trial sometime in the 1940’s and its development was dropped. The team in this study decided to go back and take a second look at nourseothricin.
“We started searching around for drugs that we could use, and it turns out these super resistant bugs were highly susceptible to streptothricin, so we were able to use it as a selection agent to do these experiments,” study co-author and pathologist at Harvard Medical School James Kirby said in a statement. “What scientists were isolating in 1942 was not as pure as what we are working with today. In fact, what was then called streptothricin is actually a mixture of several streptothricin variants. The natural mixture of different types of streptothricins is now referred to as nourseothricin.”
Kirby is also the director of the Clinical Microbiology Laboratory at Beth Israel Deaconess Medical Center.
In the earlier studies on the antibiotic, nourseothricin suffered from incomplete purification of streptothricin which was likely causing the toxicity. A study published in 2022 showed that multiple forms of streptothricin actually have different toxicities.
One called streptothricin-F was significantly less toxic while also working against present day pathogens that are resistant to multiple drugs.
In this study, the team looked closely at streptothricin-F and also streptothricin- D. Streptothricin-D strain was also highly selective for the gram-negative bacteria and was even more powerful than streptothricin-F against drug-resistant Enterobacterales and other bacterial species. However, it caused renal toxicity at a lower dose.
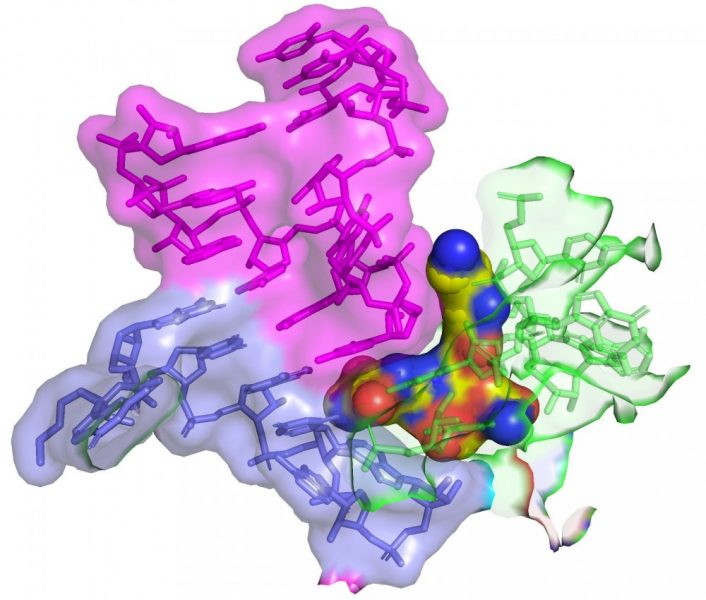
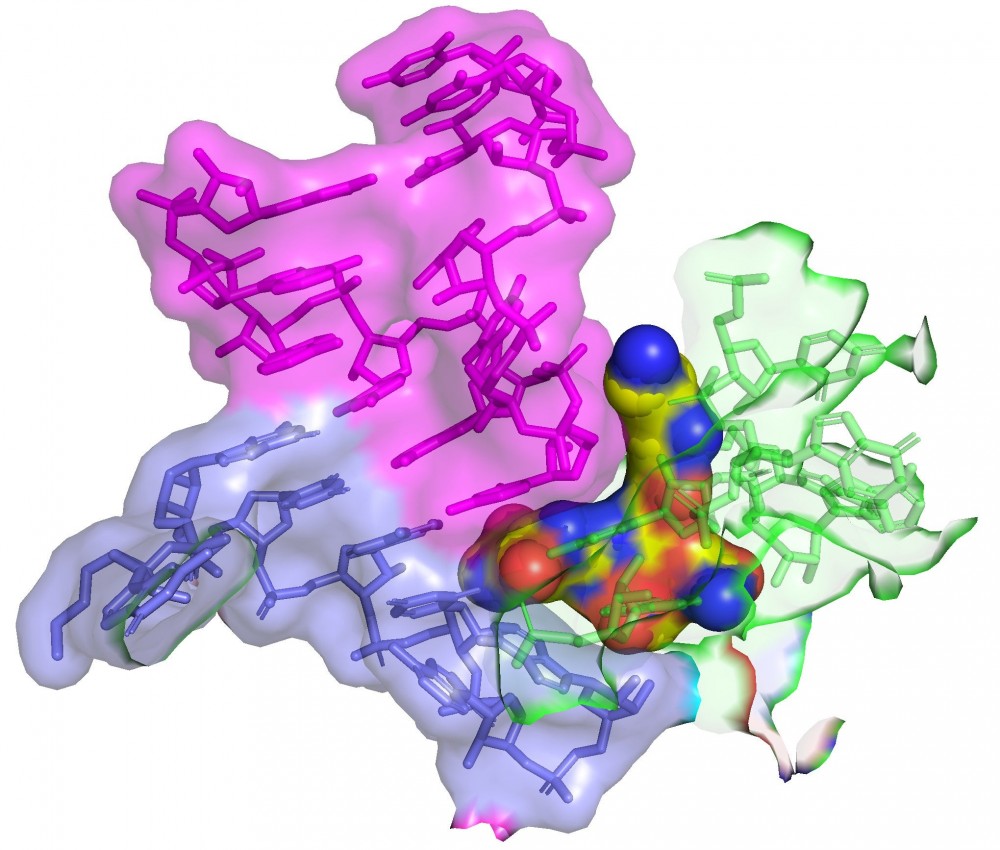
The team used cryo-electron microscopy to show that streptothricin-F bound extensively to a subunit of the bacterial ribosome. This binding causes translation errors in the bacteria, which helps antibiotics inhibit the spread of a bacterial infection.
“It works by inhibiting the ability of the organism to produce proteins in a very sneaky way. When a cell makes proteins, they make them off a blueprint or message that tells the cell what amino acids to link together to build the protein. Our studies help explain how this antibiotic confuses the machinery so that the message is read incorrectly, and it starts to put together gibberish. Essentially the cell gets poisoned because it’s producing all this junk,” said Kirby.
The team is still trying to figure out the mechanism behind how nourseothricin works, but found that it acts differently than other antibiotics. Kirby will continue studying nourseothricin with collaborators at Northwestern University and Case Western Reserve University Medical Center to dive deeper and understand how it actually works.
“We have great collaborators that have allowed us to pursue a project that crosses multiple fields. This work is an example of collaborative science really at its best,” said Kirby.